pacman::p_load(shiny, tidyverse, shinydashboard,dplyr,
spatstat, spdep,
lubridate, leaflet,
plotly, DT, viridis,
ggplot2, sf, tmap, readr,
scales, ggthemes, gridExtra,
knitr, data.table,
CGPfunctions, ggHoriPlot,
patchwork, ggiraph, vcd, vcdExtra,
ggstatsplot, ggmosaic, FunnelPlotR,
knitr)Take Home Ex 4.1 - Project Charts
Installing and Launching R Packages
Data Wrangling
Importing Dataset
Myanmar <- read_csv("data/2010-01-01-2023-12-31-Southeast_Asia-Myanmar.csv")Adjusting Attributes
Myanmar <- Myanmar %>%
mutate(year =factor(year))
Myanmar$event_date <- dmy(Myanmar$event_date)Reducing Columns
Myanmar_final <- Myanmar %>%
select(-time_precision, -geo_precision, -source_scale, -timestamp, -tags)Geo-data Correction
ACLED_MMR_1 <- Myanmar_final %>%
mutate(admin1 = case_when(
admin1 == "Bago-East" ~ "Bago (East)",
admin1 == "Bago-West" ~ "Bago (West)",
admin1 == "Shan-North" ~ "Shan (North)",
admin1 == "Shan-South" ~ "Shan (South)",
admin1 == "Shan-East" ~ "Shan (East)",
TRUE ~ as.character(admin1)
))ACLED_MMR_1 <- Myanmar_final %>%
mutate(admin2 = case_when(
admin2 == "Yangon-East" ~ "Yangon (East)",
admin2 == "Yangon-West" ~ "Yangon (West)",
admin2 == "Yangon-North" ~ "Yangon (North)",
admin2 == "Yangon-South" ~ "Yangon (South)",
admin2 == "Mong Pawk (Wa SAD)" ~ "Tachileik",
admin2 == "Nay Pyi Taw" ~ "Det Khi Na",
admin2 == "Yangon" ~ "Yangon (West)",
TRUE ~ as.character(admin2)
))EDA
Summary of Incidents
Summary_Data <- ACLED_MMR_1 %>%
group_by(year,event_type) %>%
summarize(
Total_incidents = n(),
Total_Fatalities = sum(fatalities, na.rm=TRUE)
)Distribution of Incidents Across Years by event_type
gg1 <- ggplot(Summary_Data,
aes(x = year,
y = Total_incidents,
fill = event_type)) +
geom_col_interactive(aes(tooltip = year,
data_id = year),
alpha = 0.5) +
geom_text(aes(label = Total_incidents),
vjust = 0.5,
color = "black",
size = 2,
check_overlap = TRUE,
position = "dodge") +
facet_wrap(~event_type,
ncol =2) +
theme_minimal() +
labs(y = "Total Number of Incidents", x = "Years") +
theme(
panel.grid.major.y = element_line(color = "pink", linetype = 2),
strip.background = element_rect(fill = "black"),
axis.text.x = element_blank(),
strip.text = element_text(colour = "white"),
legend.position = "none")
girafe(
ggobj = gg1,
width_svg = 6,
height_svg = 6*0.618)Distribution of Fatalities Across Years by event_type
gg2 <- ggplot(Summary_Data,
aes(x = year,
y = Total_Fatalities,
fill = event_type)) +
geom_col_interactive(aes(tooltip = year,
data_id = year),
alpha = 0.5) +
geom_text(aes(label = Total_incidents),
vjust = 0.5,
color = "black",
size = 2,
check_overlap = TRUE,
position = "dodge") +
facet_wrap(~event_type,
ncol =2) +
theme_minimal() +
labs(y = "Total Number of Fatalities", x = "Years") +
theme(
panel.grid.major.y = element_line(color = "pink", linetype = 2),
strip.background = element_rect(fill = "black"),
strip.text = element_text(colour = "white"),
axis.text.x = element_blank(),
legend.position = "none")
girafe(
ggobj = gg2,
width_svg = 6,
height_svg = 6*0.618)Distribution of Incidents by Region
Region_Summary <- ACLED_MMR_1 %>%
group_by(country, admin1, admin2, admin3, event_type,sub_event_type, disorder_type) %>%
summarize(
Total_incidents = n(),
Total_Fatalities = sum(fatalities, na.rm=TRUE)
)gg3 <- ggplot(Region_Summary,
aes(x = admin1,
y = Total_incidents,
fill = event_type)) +
geom_col_interactive(aes(tooltip = admin1,
data_id = admin1),
alpha = 0.5) +
geom_text(aes(label = Total_incidents),
vjust = 0.5,
color = "black",
size = 2,
check_overlap = TRUE,
position = "dodge") +
facet_wrap(~event_type,
ncol =2) +
theme_minimal() +
labs(y = "Total Number of Incidents", x = "Regions") +
theme(
panel.grid.major.y = element_line(color = "pink", linetype = 2),
axis.text.x = element_blank(),
strip.background = element_rect(fill = "black"),
strip.text = element_text(colour = "white"),
legend.position = "none")
girafe(
ggobj = gg3,
width_svg = 6,
height_svg = 6*0.618)Distribution of Fatalities by Region
gg4 <- ggplot(Region_Summary,
aes(x = admin1,
y = Total_Fatalities,
fill = event_type)) +
geom_col_interactive(aes(tooltip = admin1,
data_id = admin1),
alpha = 0.5) +
geom_text(aes(label = Total_Fatalities),
vjust = 0.5,
color = "black",
size = 2,
check_overlap = TRUE,
position = "dodge") +
facet_wrap(~event_type,
ncol =2) +
theme_minimal() +
labs(y = "Total Number of Fatalities", x = "Regions") +
theme(
panel.grid.major.y = element_line(color = "pink", linetype = 2),
axis.text.x = element_blank(),
strip.background = element_rect(fill = "black"),
strip.text = element_text(colour = "white"),
legend.position = "none")
girafe(
ggobj = gg4,
width_svg = 6,
height_svg = 6*0.618)CDA
Summary of Event Types by Fatalities (Total Count)
ggbetweenstats(ACLED_MMR_1,
x= event_type,
y= fatalities)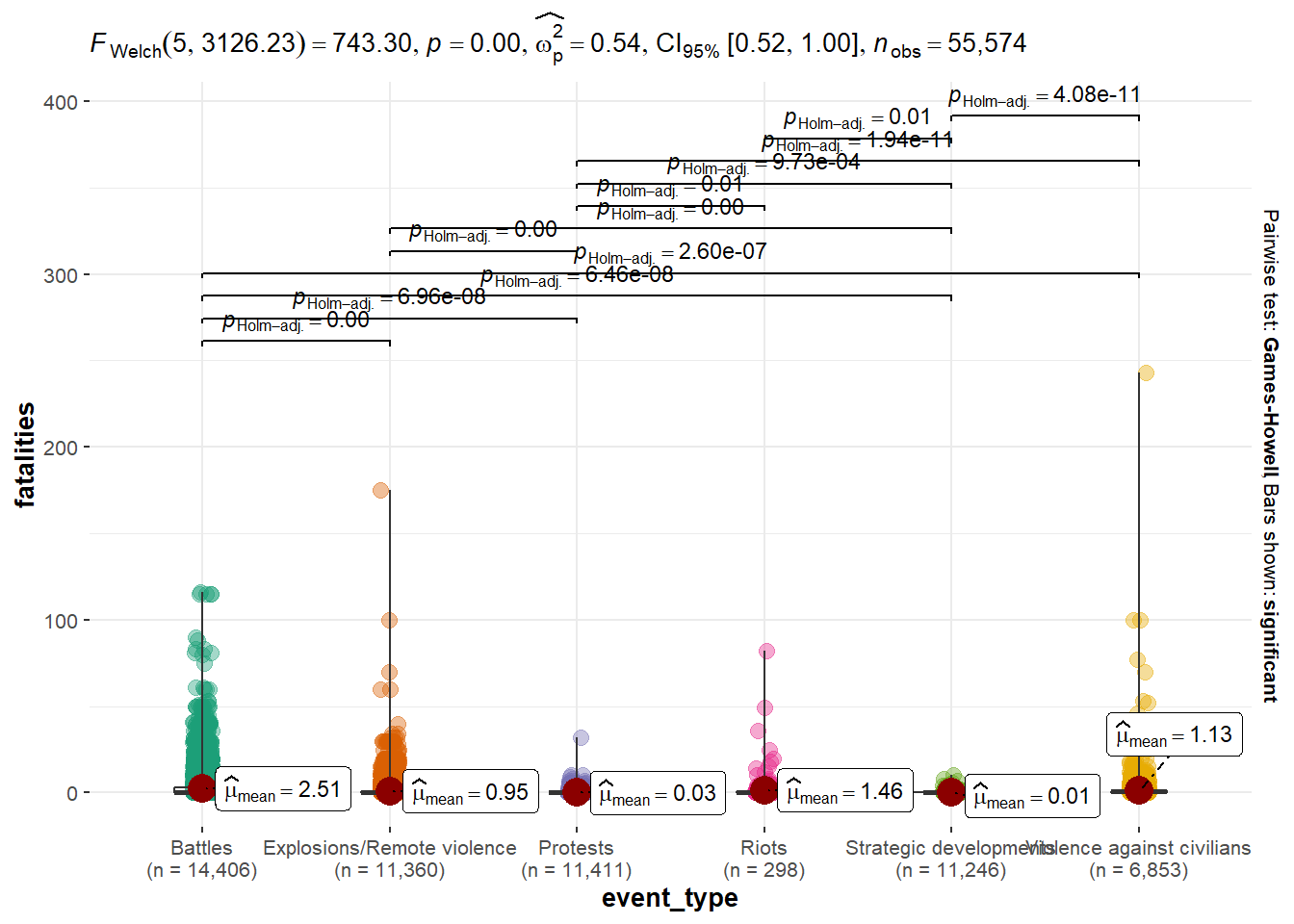
Summary of Event Types by Fatalities (Total Count), Grouped by Region
grouped_ggbetweenstats(ACLED_MMR_1,
x = event_type,
y = fatalities,
grouping.var = admin1,
type = "np",
pairwise.display = "s",
pairwise_comparisons = TRUE,
output = "plot")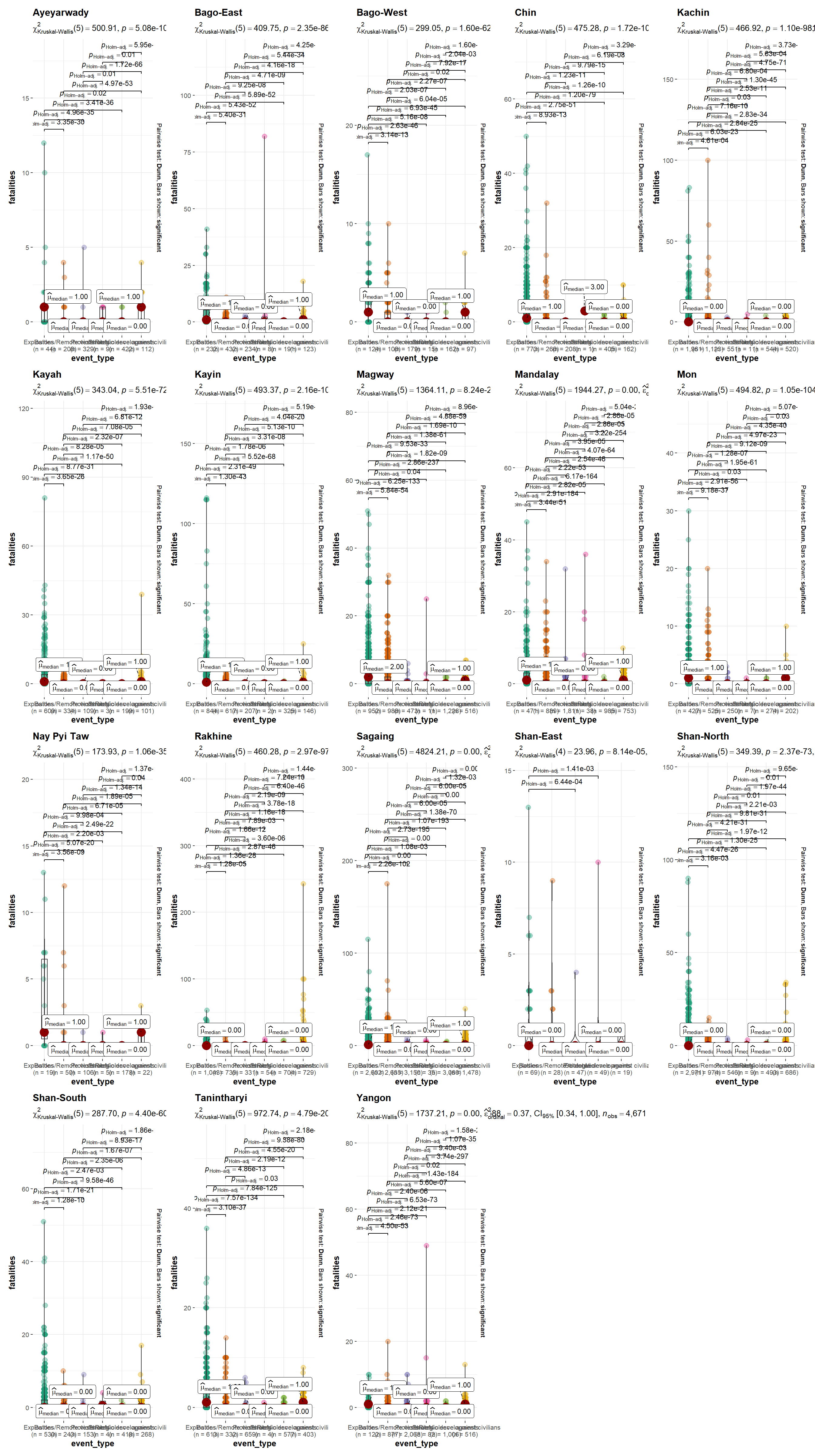
Summary of Event Types Across Years, Incidents and Fatalities
Incidents
ggbetweenstats(Summary_Data,
x= event_type,
y= Total_incidents)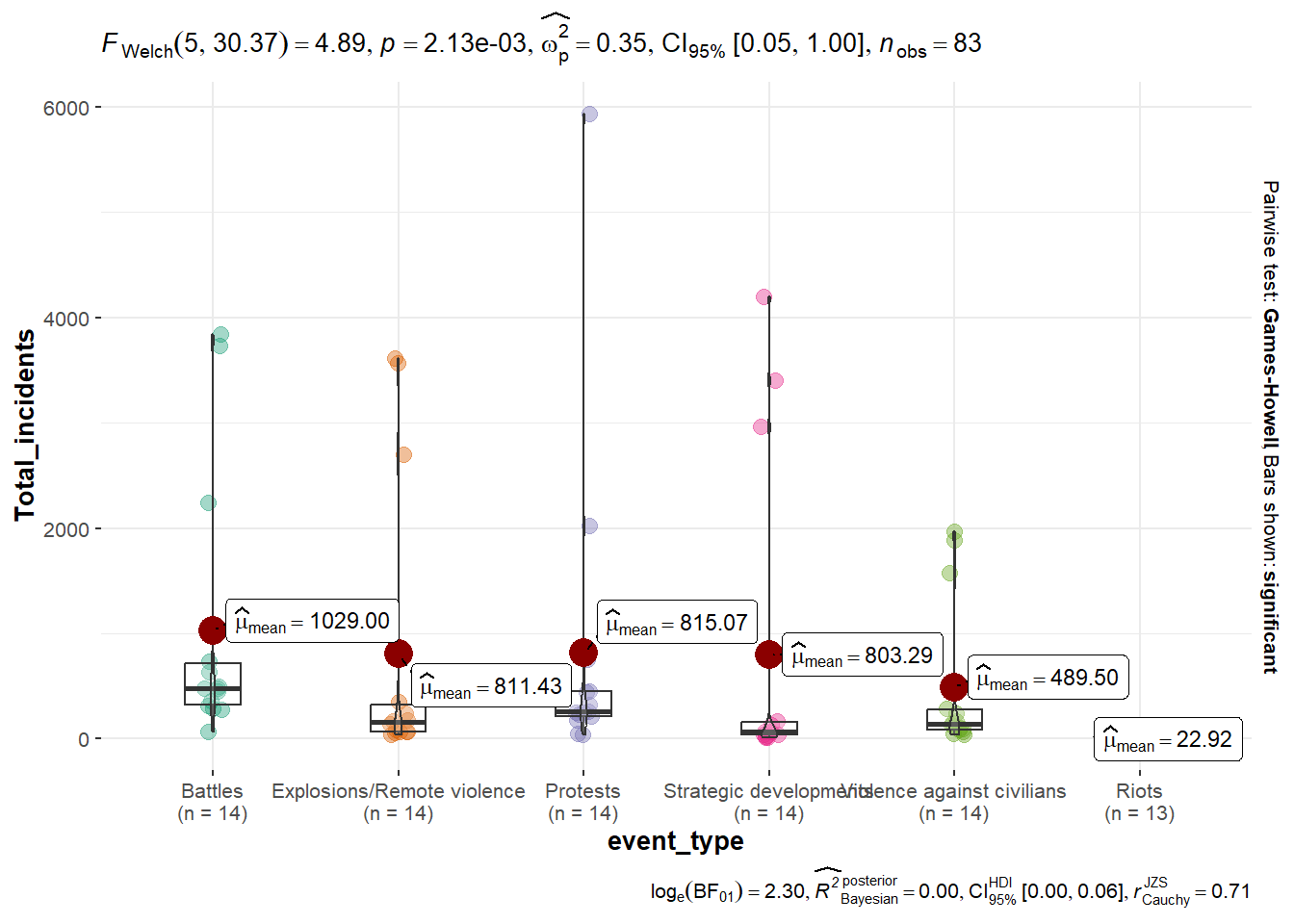
Fatalities
ggbetweenstats(Summary_Data,
x= event_type,
y= Total_Fatalities)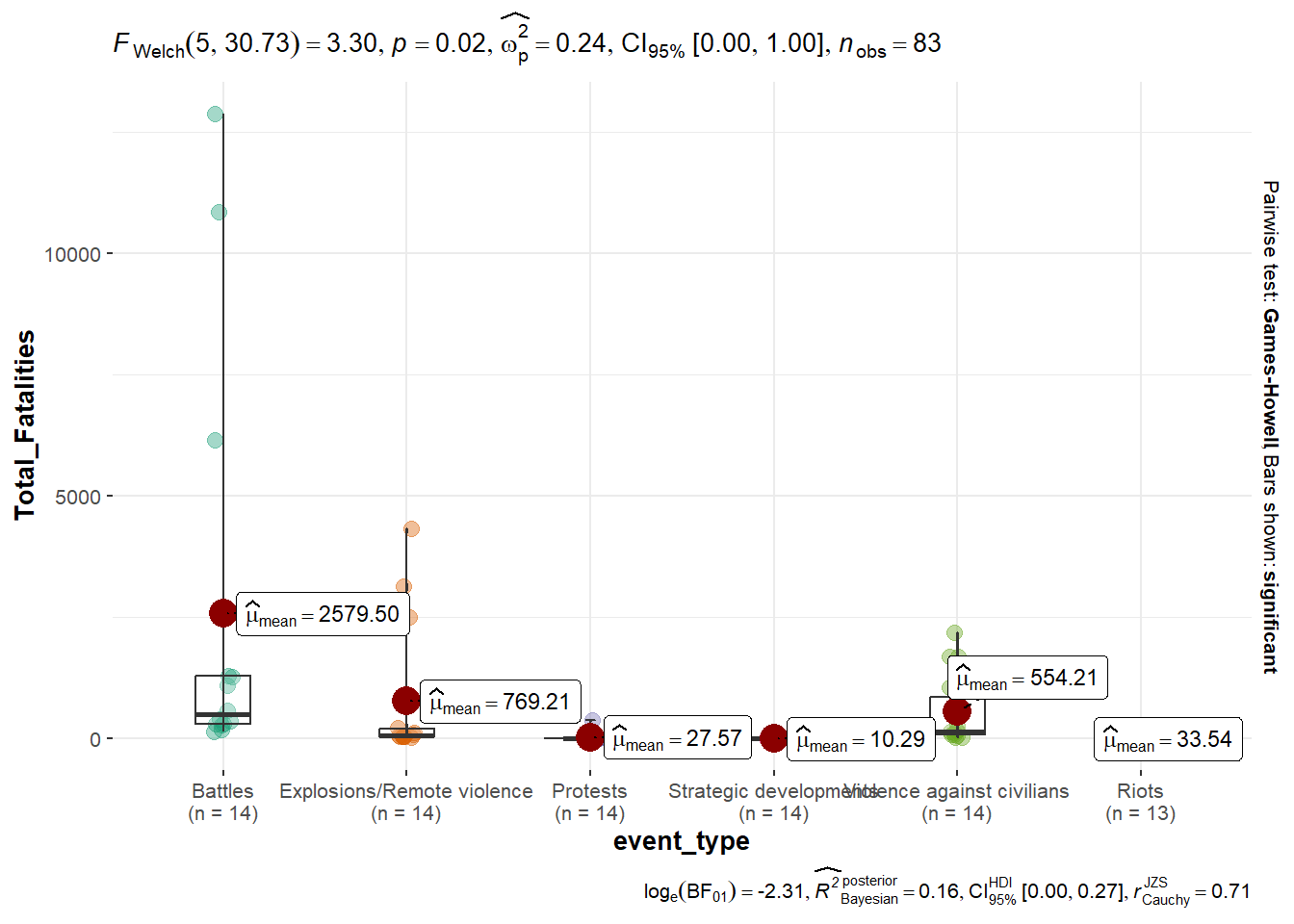
Mosaic Plot (Incident Counts)
By Year and Event Type
vcd::mosaic(~year + event_type, data = ACLED_MMR_1, gp = shading_max,
labeling = labeling_border(rot_labels = c(90,0,0,0),
just_labels = c("left",
"center",
"center",
"right")))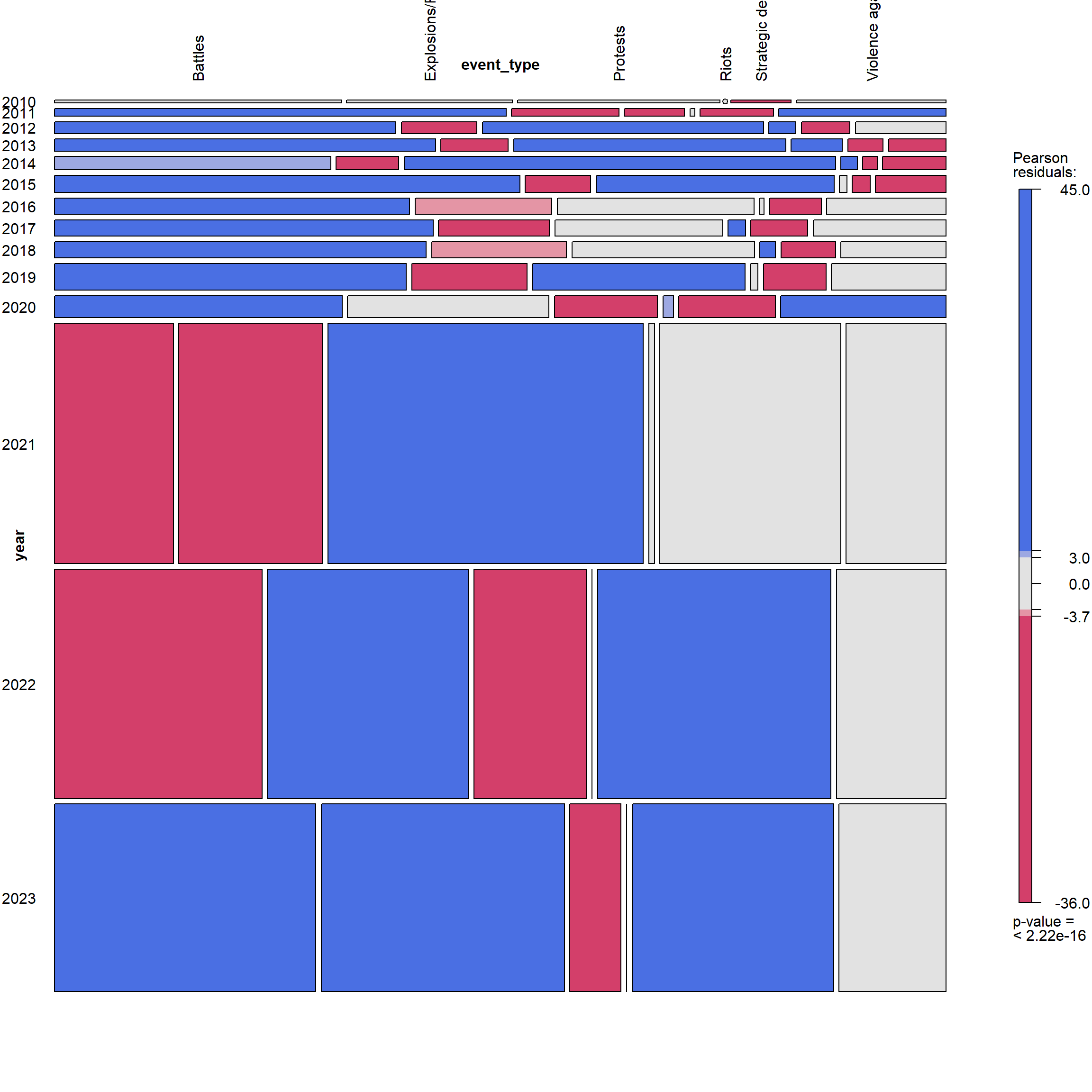
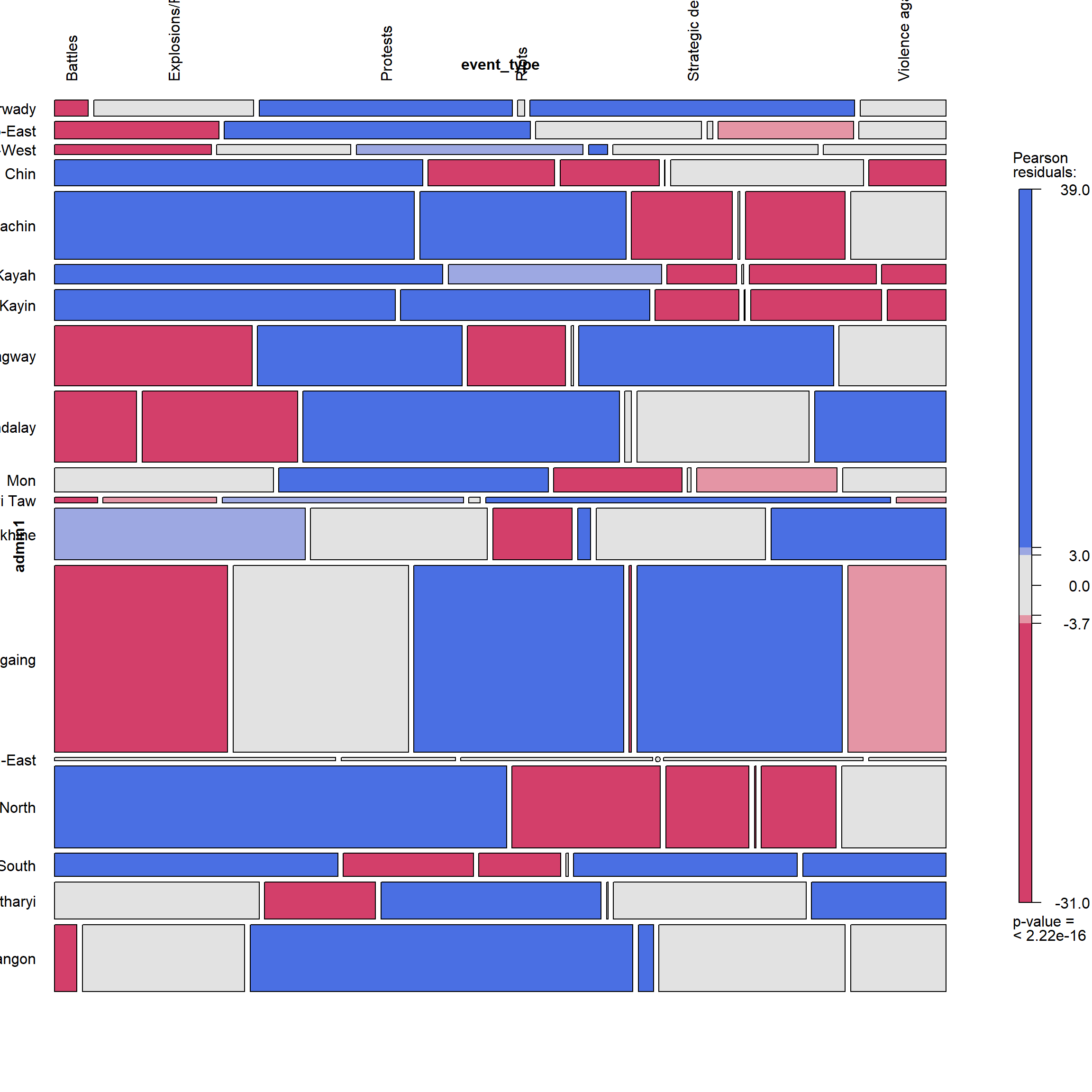
By Region and Event Type
vcd::mosaic(~ admin1 + event_type, data = ACLED_MMR_1, gp = shading_max,
labeling = labeling_border(rot_labels = c(90,0,0,0),
just_labels = c("left",
"center",
"center",
"right")
))
admin1 + disorder_type
vcd::mosaic(~ admin1 + disorder_type, zero_size = FALSE, data = ACLED_MMR_1,gp = shading_max,
labeling = labeling_border(rot_labels = c(90,0,0,0),
just_labels = c("left",
"center",
"center",
"right")))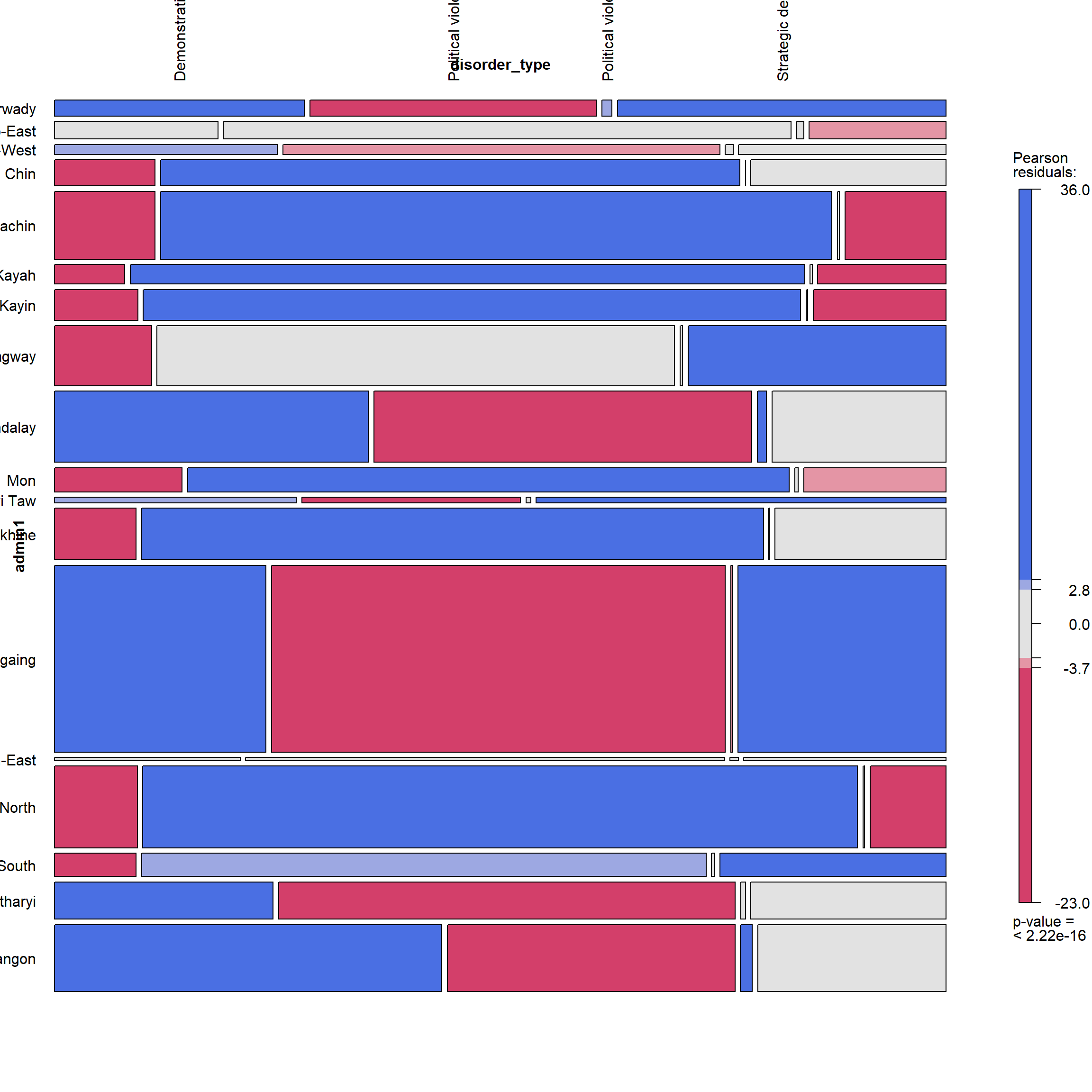
subeventtype
vcd::mosaic(~ sub_event_type +admin1, zero_size = FALSE, data = ACLED_MMR_1,gp = shading_max,
labeling = labeling_border(rot_labels = c(90,0,0,0),
just_labels = c("left",
"center",
"center",
"right")))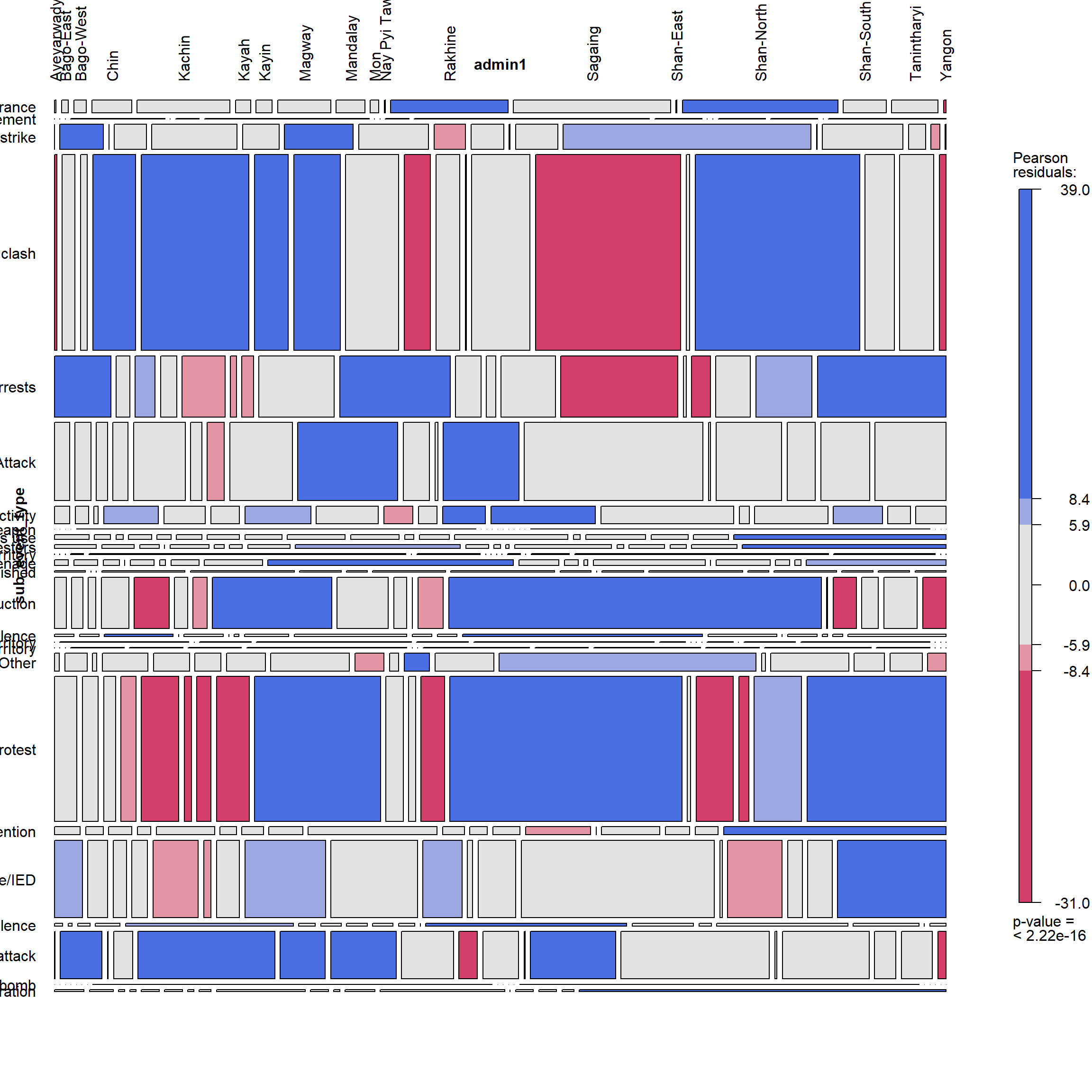
Using geom_mosiac
Fatalities by Event Type and Region
gg5 <- ggplot(Region_Summary) +
geom_mosaic(aes(weight = Total_Fatalities,
x = (product(event_type, country)), fill = admin1)) +
labs(x = "Myanmar",
fill = "Regions") +
## guides(fill = "none") + to remove legend if not required
theme(
axis.text.x = element_blank(),
axis.title.y = element_blank(),
axis.ticks.x = element_blank()
)
ggplotly(gg5)gg6 <- ggplot(Region_Summary) +
geom_mosaic(aes(weight = Total_Fatalities,
x = (product(sub_event_type, country)), fill = admin2)) +
labs(x = "Myanmar",
fill = "Territory") +
## guides(fill = "none") + to remove legend if not required
theme(
axis.text.x = element_blank(),
axis.title.y = element_blank(),
axis.ticks.x = element_blank()
)
ggplotly(gg6)FunnelPlotR
Region_Summary_Test <- ACLED_MMR_1 %>%
filter(year == 2023)
Region_Summary_Test <- Region_Summary_Test %>%
group_by(country, admin1, admin2, admin3, event_type) %>%
summarize(
Total_incidents = n(),
Total_fatalities = sum(fatalities)
)
Region_Summary_Test <- Region_Summary_Test %>%
mutate(rate = Total_fatalities / Total_incidents) %>%
mutate(rate.se = sqrt((rate*(1-rate))/ (Total_incidents))) %>%
filter(rate>0)fit.mean <- weighted.mean(Region_Summary_Test$rate, 1/Region_Summary_Test$rate.se^2)number.seq <- seq(1, max(Region_Summary_Test$Total_incidents), 1)
number.ll95 <- fit.mean - 1.96 * sqrt((fit.mean*(1-fit.mean)) / (number.seq))
number.ul95 <- fit.mean + 1.96 * sqrt((fit.mean*(1-fit.mean)) / (number.seq))
number.ll999 <- fit.mean - 3.29 * sqrt((fit.mean*(1-fit.mean)) / (number.seq))
number.ul999 <- fit.mean + 3.29 * sqrt((fit.mean*(1-fit.mean)) / (number.seq))
dfCI <- data.frame(number.ll95, number.ul95, number.ll999,
number.ul999, number.seq, fit.mean)p <- ggplot(Region_Summary_Test, aes(x = Total_incidents, y = rate)) +
geom_point(alpha=0.4) +
geom_line(data = dfCI,
aes(x = number.seq,
y = number.ll95),
size = 0.4,
colour = "grey40",
linetype = "dashed") +
geom_line(data = dfCI,
aes(x = number.seq,
y = number.ul95),
size = 0.4,
colour = "grey40",
linetype = "dashed") +
geom_line(data = dfCI,
aes(x = number.seq,
y = number.ll999),
size = 0.4,
colour = "grey40") +
geom_line(data = dfCI,
aes(x = number.seq,
y = number.ul999),
size = 0.4,
colour = "grey40") +
geom_hline(data = dfCI,
aes(yintercept = fit.mean),
size = 0.4,
colour = "grey40")
p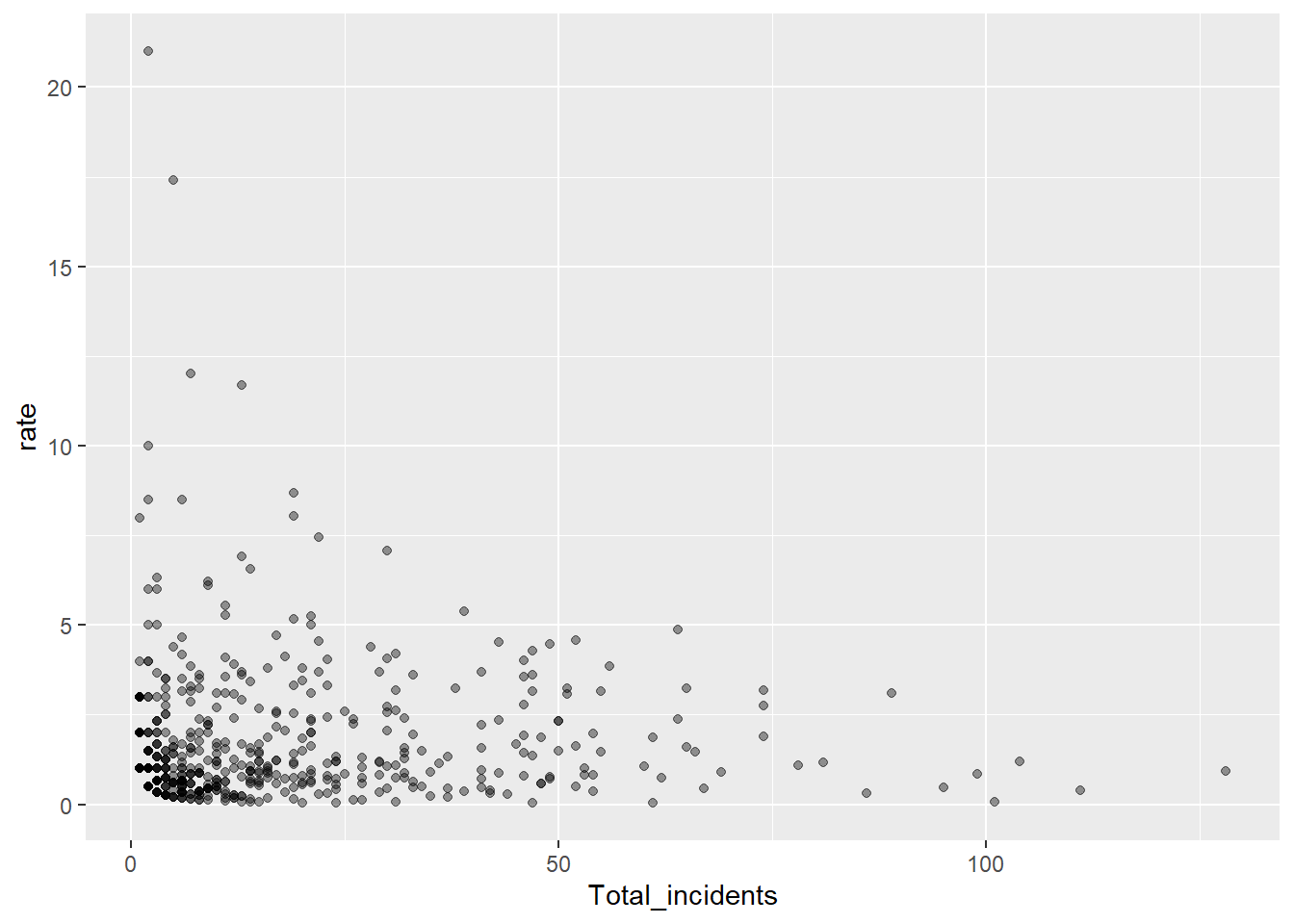
funnel_plot(
numerator = Region_Summary_Test$Total_incidents,
denominator = Region_Summary_Test$Total_fatalities,
group = Region_Summary_Test$admin2,
data_type = "PR", #<<
xrange = c(0,1500), #<<
yrange = c(0,20) #<<
)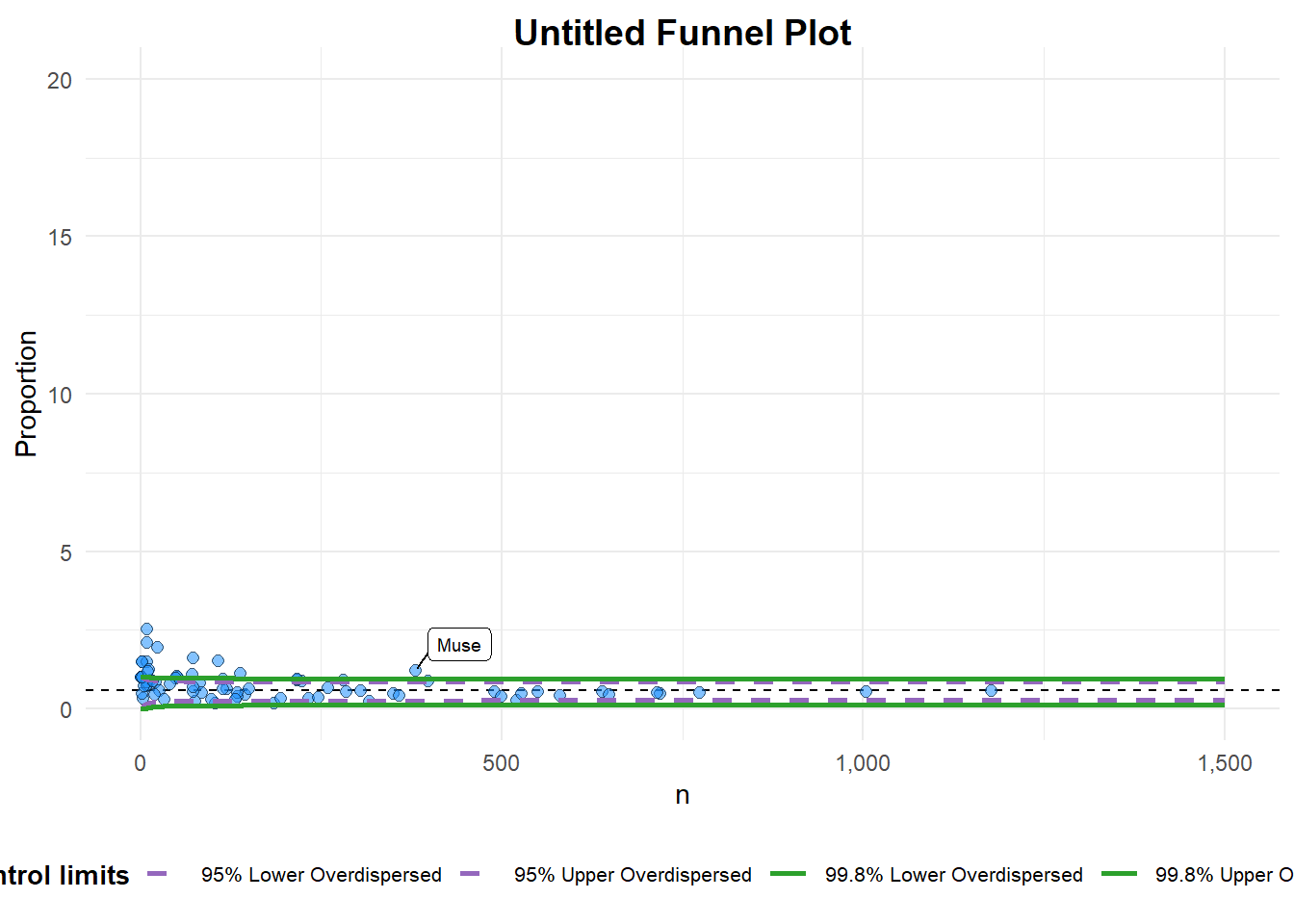
A funnel plot object with 76 points of which 16 are outliers.
Plot is adjusted for overdispersion.